title: 'GeneNetwork: framework for web-based genetics' tags: - bioinformatics - genetics - genomics authors: - name: Zachary Sloan orcid: 0000-0002-8099-1363 affiliation: University of Tennessee Health Science Center, USA - name: Danny Arends orcid: 0000-0001-8738-0162 affiliation: Humboldt University, Berlin, Germany - name: Karl W. Broman orcid: 0000-0002-4914-6671 affiliation: University of Wisconsin, USA - name: Arthur Centeno orcid: 0000-0003-3142-2081 affiliation: University of Tennessee Health Science Center, USA - name: Nicholas Furlotte orcid: 0000-0002-9096-6276 - name: Harm Nijveen orcid: 0000-0002-9167-4945 affiliation: Wageningen University, The Netherlands - name: Lei Yan orcid: 0000-0001-5259-3379 affiliation: University of Tennessee Health Science Center, USA - name: Xiang Zhou orcid: 0000-0002-4331-7599 affiliation: University of Michigan - name: Robert W. Williams orcid: 0000-0001-8924-4447 affiliation: University of Tennessee Health Science Center, USA - name: Pjotr Prins orcid: 0000-0002-8021-9162 affiliation: University Medical Center Utrecht, The Netherlands, University of Tennessee Health Science Center, USA date: 29 May 2016 bibliography: paper.bib
Summary
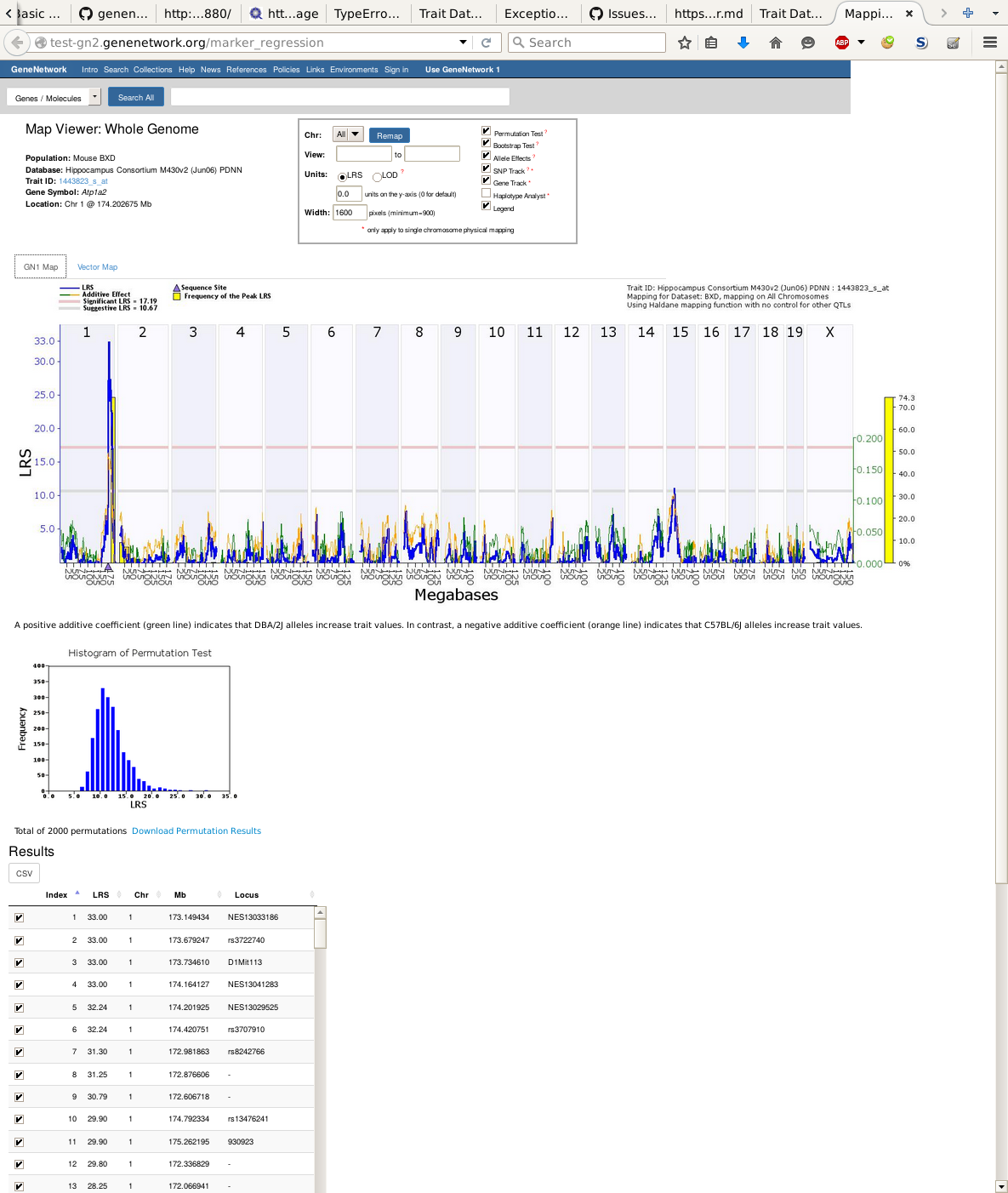
GeneNetwork (GN) is a free and open source (FOSS) framework for web-based genetics that can be deployed anywhere. GN allows biologists to upload high-throughput experimental data, such as expression data from microarrays and RNA-seq, and also `classic' phenotypes, such as disease phenotypes. These phenotypes can be mapped interactively against genotypes using embedded tools, such as R/QTL [@Arends:2010] mapping, interval mapping for model organisms and pylmm; an implementation of FaST-LMM [@Lippert:2011] which is more suitable for human populations and outbred crosses, such as the mouse diversity outcross. Interactive D3 graphics are included from R/qtlcharts and presentation-ready figures can be generated. Recently we have added functionality for phenotype correlation [@Wang:2016] and network analysis [@WGCNA:2008].
-
GN is written in python and javascript and contains a rich set of tools and libraries that can be written in any computer language. A full list of included software can be found in the package named `genenetwork2' and defined in guix-bioinformatics. To make it easy to install GN locally in a byte reproducible way, including all dependencies and a 2GB MySQL test database (the full database is 160GB and growing), GN is packaged with GNU Guix, as described here. GNU Guix deployment makes it feasible to deploy and rebrand GN anywhere.
Future work
More mapping tools will be added, including support for Genome-wide Efficient Mixed Model Association (GEMMA). The Biodiallance genome browser is being added as a Google Summer of Code project with special tracks related to QTL mapping and network analysis. Faster LMM solutions are being worked on, including GPU support.
A REST interface is being added so that data can be uploaded to a server, analysis run remotely on high performance hardware, and results downloaded and used for further analysis. This feature will allow biologist-programmers to use R and Python on their computer and execute computations on GN enabled servers.
