# Storyboard: starting from a known gene
GeneNetwork contains data on thousands of experiments. How do you mine GN when you have the name of a gene? Start with [BRCA2](https://genenetwork.org/genes/BRCA2).
# Table of contents
1. [Global search](#gsearch)
1. [Gene aliases](#alias)
1. [Genewiki](#genewiki)
1. [TODO](#TODO)
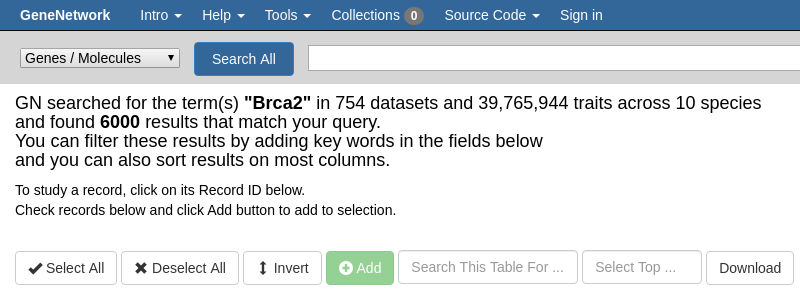
## Use the search page
The top bar of GeneNetwork shows a global search.

We can look for the `Brca2` mouse gene:
## Find the right alias/name
GeneNetwork knows about aliases. But sometimes you want to check what aliases there are in mouse. If you search for BRCA2 and click on a search result, for example for [1448542_at](http://genenetwork.org/show_trait?trait_id=1448542_at&dataset=HC_M2_0606_P),
you get to the 'trait page' which shows the aliases:
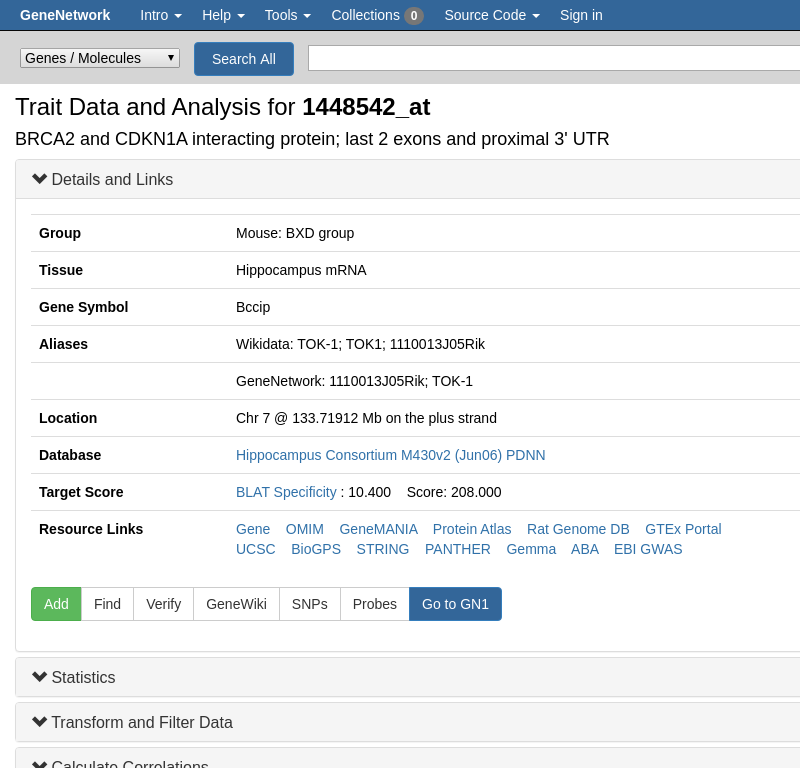
GeneNetwork ignores case by default. In a search use quotes to search case sensitive. For example, [BRCA2](https://www.wikidata.org/wiki/Q17853272) on wikidata.org refers to the human gene. The mouse gene is named [Brca2](https://www.wikidata.org/wiki/Q14864738).
## Find the GeneNetwork genewiki entry
GeneNetwork maintains info on gene entries. For example for [BRCA2](http://gn1.genenetwork.org/webqtl/main.py?FormID=geneWiki&symbol=BRCA2).
## TODO
* [ ] expand story line
* [ ] global search is faulty, see gn-gemtext-threads/issues/genenetwork/global-search.gmi
* [ ] migrate GeneWiki entries


